Usecase ea: Difference between revisions
From referenceTSS
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
{{DISPLAYTITLE: }} | {{DISPLAYTITLE: }} | ||
=== <center> | === <center> Enrichment Analyses </center> === | ||
---- | ---- | ||
One of the standard approaches in downstream analysis of gene expression is enrichment analysis that can be useful for evaluating biological function under condition. In this use-case, two scenarios of annotation enrichment analyses are introduced for refTSS users. | One of the standard approaches in downstream analysis of gene expression is enrichment analysis that can be useful for evaluating biological function under condition. In this use-case, two scenarios of annotation enrichment analyses are introduced for refTSS users. | ||
* ''' | * ''' Gene enrichment analyses''' | ||
: '''GO and KEGG pathway annotation enrichment analysis''' are commonly used for examination of the over-representation of gene annotations. Here we introduce the workflow of general annotation enrichment analysis using refTSSs. | : '''GO and KEGG pathway annotation enrichment analysis''' are commonly used for examination of the over-representation of gene annotations. Here we introduce the workflow of general annotation enrichment analysis using refTSSs. | ||
| Line 21: | Line 21: | ||
</html> | </html> | ||
==== 1. | ==== 1. Gene annotation enrichment analysis ==== | ||
: Annotation enrichment analysis examines the over-representation of genes of interest with biological annotations such as GO and KEGG pathways. To apply TSS analysis using refTSS to | : Annotation enrichment analysis examines the over-representation of genes of interest with biological annotations such as GO and KEGG pathways. To apply TSS analysis using refTSS to gene annotation enrichment, '''TSS IDs must be converted to gene IDs or symbols ''' for input into annotation enrichment tools. refTSS4 has newly launched a simple web tool for converting TSS IDs to gene symbols to support refTSS users. | ||
<html> | <html> | ||
| Line 37: | Line 37: | ||
</html> | </html> | ||
1.3. '''External enrichment tools (e.g., DAVID, Metascape, TopGO).''' | 1.3. '''External enrichment tools (e.g., DAVID, Metascape, TopGO).''' | ||
: | <li> [[Source::https://david.ncifcrf.gov|DAVID]] </li> | ||
: # Metascape | <li> [[Source::https://metascape.org/gp/index.html#/main/step1|Metascape]] </li> | ||
: | <li> [[Source::https://bioconductor.org/packages/release/bioc/html/topGO.html|TopGO (R package)]] </li> | ||
: | <li> [[Source::https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html|ClusterProfiler (R package)]] </li> | ||
: Metascape | |||
: TopGO (R package) | |||
: ClusterProfiler (R package) | |||
:: The extracted Gene Symbols are input into external enrichment analysis tools such as '''DAVID, MetaScape, and ClusterProfiler'''. | :: The extracted Gene Symbols are input into external enrichment analysis tools such as '''DAVID, MetaScape, and ClusterProfiler'''. | ||
Revision as of 11:56, 6 November 2024
Enrichment Analyses
One of the standard approaches in downstream analysis of gene expression is enrichment analysis that can be useful for evaluating biological function under condition. In this use-case, two scenarios of annotation enrichment analyses are introduced for refTSS users.
- Gene enrichment analyses
- GO and KEGG pathway annotation enrichment analysis are commonly used for examination of the over-representation of gene annotations. Here we introduce the workflow of general annotation enrichment analysis using refTSSs.
- GWAS-LD enrichment analysis
- Novel enrichment service called refTSS-LD was lunched on refTSS4. refTSS-LD provides GWAS-LD enrichment analysis using refTSS IDs.
For checking both approaches, there is an example input file of TSS ID lists from CAGE analysis of neural crest cell differentiation experiments with iPS cells (day0_vs_day18).
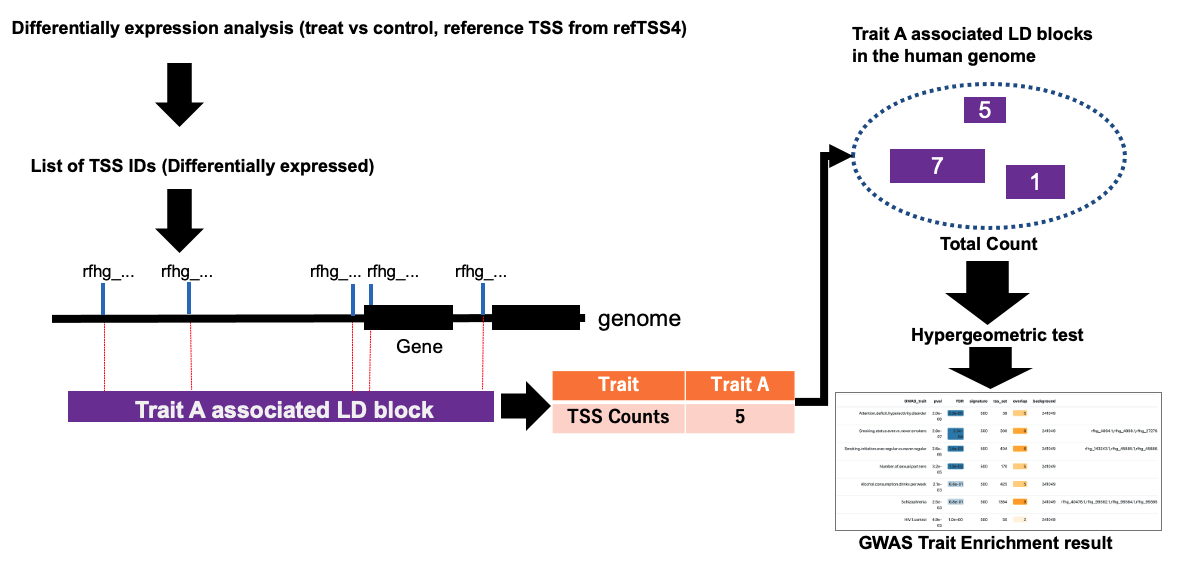
1. Gene annotation enrichment analysis
- Annotation enrichment analysis examines the over-representation of genes of interest with biological annotations such as GO and KEGG pathways. To apply TSS analysis using refTSS to gene annotation enrichment, TSS IDs must be converted to gene IDs or symbols for input into annotation enrichment tools. refTSS4 has newly launched a simple web tool for converting TSS IDs to gene symbols to support refTSS users.
1.1. TSS expression analysis and extraction of statistically significant TSSIDs.
- TSS sequencing is useful method for evaluation of gene expression value using TSS tags. refTSS4 introduces how to perform differential analysis of TSS on Tutorial of TSS differential analysis .
1.2. ID conversion from TSSID to Official Gene symbols.
- Conversion of differentially expressed TSSID to ID conversion tools
-
1.3. External enrichment tools (e.g., DAVID, Metascape, TopGO).
- Metascape
- TopGO (R package)
- ClusterProfiler (R package)
- The extracted Gene Symbols are input into external enrichment analysis tools such as DAVID, MetaScape, and ClusterProfiler.
2. GWAS-LD enrichment analysis on refTSS-LD
- As well as the other enrichment tools, refTSS provides simple enrichment analysis service, refTSS-LD. This service provides GWAS-LD enrichment analysis.
-
- From the menu, select refTSS-LD and paste the TSS IDs into the box.
-
- The table shows the results of the GWAS-LD analysis conducted using the example data. Results are sorted by order of FDR. The enriched TSS IDs are displayed in the "hits" column, allowing for inspection of which TSSs are concentrated in LD blocks grouped by traits.